I’ve created an R Package (photoforestryr) meant to help marine biologists to analyze forestry measurements in mangroves. Unlike “terrestrial” forests, mangroves are extremely difficult to traverse given the tangle of aerial and prop roots that are characteristic of Rhizophora spp. (Figure 1). This tangle of structure makes the task of collecting tree density, diameter at breast height (DBH), basal area, and absolute height measurements prohibitively challenging; what may take minutes in a pine forest will take hours in mangroves.
Figure 1. Mangrove understory.
A solution is to adopt more indirect methods that do not require the observer to physically contact each tree that is to be measured. By using a combination of plotless density estimation methods, trigonometry and photogrammetric interpretation, I found a way to describe whole mangrove stands from a fixed point rather than needing to move from tree to tree.
To perform a survey using this method, the observer sets up a tripod at a random point within the forest. Tree Density can be determined using a laser range finder and the Point-centered Quarter method (Figure 2). The diameter of each tree at 1.3 m above the ground (DBH) can be accurately estimated using an empirically established relationship between pixel number and distance for a given camera. This relationship was found by photographing a post of known diameter from various distances and observing the number of pixels (using ImageJ image analysis software) occupied by the post’s width at each distance (Figure 3). A regression revealed a negative exponential relationship. The exact mathematical relationship between pixel number and distance will vary from camera to camera.
Figure 2. Fixed point sampling in a mangrove forest using a camera and laser range finder.

Figure 3. Relationship between pixel number and distance for an object of known diameter.
The diameter at breast height of each tree can be calculated using the function PhotoDBH which is included in the package photoforestryr:
PhotoDBH <- function(pixels, distx){
DBH <- pixels/(33.198*(distx^-1.018));
return(DBH);
}
The resulting DBH can be used by another function included in this package, called BasalArea:
BasalArea <-function(DBH){
BA <- pi*((DBH/2)^2);
return(BA);
}
When these functions are applied to the 25 trees closest to the fixed observation point, the area -standardized basal area of the mangrove can be estimated. Basal Area is a standard estimate of standing biomass in forests and permits comparison between forests.
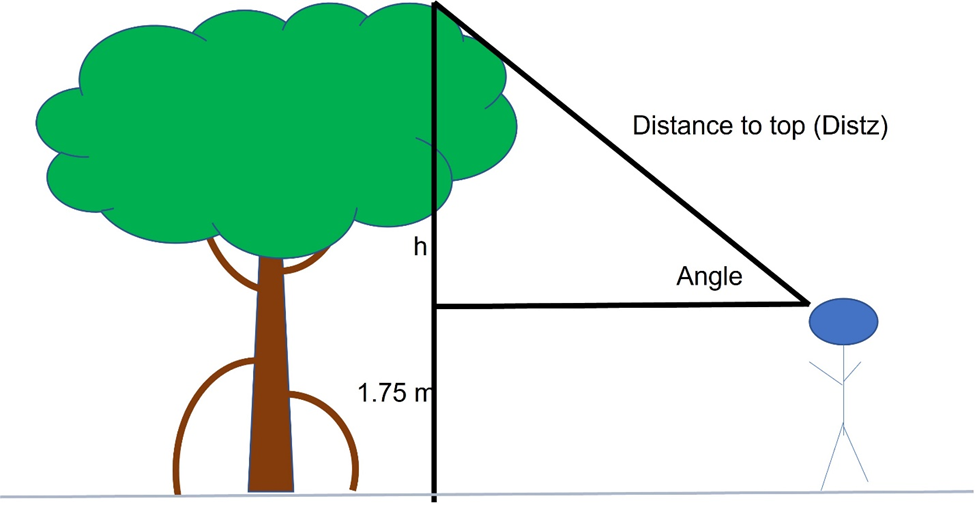
It is also possible to estimate the height of each tree observed from a fixed point using the laser range finder, a clinometer, and trigonometry. A clinometer is a handheld device that permits an observer to measure the angle between an observer and the top of a tree (Figure 4).
Figure 4. Height determination using range finder,
clinometer, and trigonometry.
By using this angle and the distance between the observer’s eye and the top of the tree, the height of the tree (H) can be determined using trigonometry. The function TreeHeight performs this calculation and adds 1.75 m to account for the height of the observer. This amount can be adjusted for individuals of a different height.
TreeHeight <- function(distz, angle){
radians=angle*(pi/180);
Height <- (sin(radians)*distz)+1.75 ;
return(Height);
}
This package is distributed in Github under a MIT license. It may be found here: https://github.com/Wellis11/photoforestr